Computer vision system analyses cells in microscopy videos
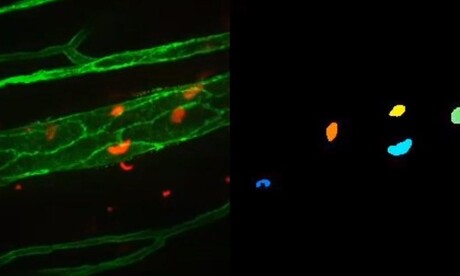
An international team of researchers, led by the Universidad Carlos III de Madrid (UC3M), has developed a system based on computer vision techniques that allows automatic analysis of biomedical videos captured by microscopy in order to characterise and describe the behaviour of the cells that appear in the images. The system incorporates machine learning as well as other types of statistical techniques and geometric models, all of which are described in the journal Medical Image Analysis.
Professor Fernando Díaz de María, Head of the UC3M Multimedia Processing Group, said his group’s contribution “consists of the design and development of a fully automatic system, based on computer vision techniques, which allows us to characterise the cells under study by analysing videos captured by biologists using the intravital microscopy technique”. Automatic measurements of the shape, size, movement and position relative to the blood vessel of a few thousand cells have been made, compared to traditional biological studies that are usually supported by analyses of a few hundred manually characterised cells. In this way, it has been possible to carry out a more advanced biological analysis with greater statistical significance.
The new system has several advantages, according to the researchers, in terms of time and precision. As explained by Ivan González Díaz, Associate Professor in the Signal Theory and Communications Department at UC3M, “It is not feasible to keep an expert biologist segmenting and tracking cells on video for months. On the other hand, to provide an approximate idea (because it depends on the number of cells and 3D volume depth), our system only takes 15 minutes to analyse a five-minute video.”
The techniques developed by the UC3M engineering team have already been used for measurements on living tissues, in research carried out with scientists from Spain’s National Centre for Cardiovascular Research (CNIC). As a result, the team discovered that neutrophils (a type of immune cell) show different behaviours in the blood during inflammatory processes and have identified that one of them, caused by the Fgr molecule, is associated with the development of cardiovascular disease. This work, published in the journal Nature, could allow the development of new treatments to minimise the consequences of heart attacks.
Deep neural networks, the tools that engineers rely on for cell segmentation and detection, are basically algorithms that learn from examples, so in order to deploy the system in a new context, it is necessary to generate sufficient examples to enable their training. That said, the software that implements the system is versatile and can be adapted to other problems in a few weeks.
“In fact,” said UC3M researcher Miguel Molina Moreno, “we are already applying it in other different scenarios, studying the immunological behaviour of T cells and dendritic cells in cancerous tissues. And the provisional results are promising.”
Please follow us and share on Twitter and Facebook. You can also subscribe for FREE to our weekly newsletters and bimonthly magazine.
AI spots hidden objects in chest scans
A new artificial intelligence (AI) tool can spot hard-to-see objects lodged in patients'...
Drying biofluid droplets used to diagnose disease
A newly developed workflow relies on the drying process of biofluid droplets to distinguish...
Map of the developing brain opens pathways to treat Parkinson's
The comprehensive single-cell map of the developing human brain captures nearly every cell type,...





