New gene editing tool could improve on CRISPR
Scientists from Columbia University Irving Medical Center have captured the first images of a new gene editing tool that could improve on existing CRISPR-based tools. Their work has been published in the journal Nature.
Many researchers around the world use CRISPR-Cas9 to quickly and cheaply make precise modifications to the genome of a cell. However, most uses of CRISPR involve cutting both strands of the target DNA, and the DNA break must then be repaired by the host cell’s own machinery.
Controlling this repair process is still a major challenge in the field, and undesired gene edits are often introduced inadvertently in the genome. Additionally, existing tools often perform poorly at inserting large genetic payloads in a precise fashion. Improving the accuracy of gene editing is thus a priority for researchers and is critical for ensuring the safety of therapies developed with this technique.
The newly developed ‘INTEGRATE’ system, on the other hand, can accurately insert large DNA sequences without relying on the cell’s machinery to repair the strands. As a result, INTEGRATE could prove to be a more accurate and efficient way of making certain gene modifications than the original CRISPR-Cas system. The new tool could also help scientists perform gene editing in cell types with limited DNA repair activity such as neurons, where attempts to use CRISPR have been comparatively less successful.
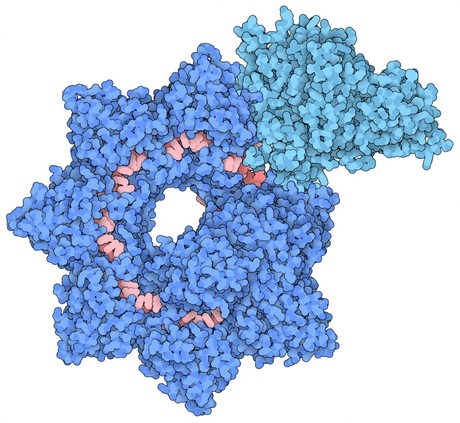
INTEGRATE was made possible after Columbia researchers, led by Assistant Professors Sam Sternberg and Israel Fernandez, discovered a ‘jumping gene’ in Vibrio cholerae bacteria that could insert large genetic payloads in the genome without introducing DNA breaks. The researchers harnessed cryo-electron microscopy to flash freeze a sample of the gene editing complex in liquid nitrogen, before bombarding it with electrons. They then used the images they captured with the electron microscope to generate atomic resolution models of the INTEGRATE system.
Their structural model reveals that the complex is made up of two main sections that are arranged in a helical filament. The larger portion, called Cascade, winds around and carries a guide RNA that it uses to scan the cell for a matching sequence in DNA. Once it locates and binds the target sequence, it threads the DNA strand through the TniQ ‘transposition’ proteins that sit on the end of the complex and recruit other enzymes that help modify the DNA.
The scanning mechanism of INTEGRATE appears to work in a similar way to other CRISPR systems, some of which also contain a Cascade complex with guide RNA. However, unlike other CRISPR systems that use Cascade to target DNA for cutting, the function of Cascade within INTEGRATE is to target DNA for highly accurate insertion of genetic payloads.
In a previous study, Asst Prof Sternberg and colleagues used genetics and biochemistry to propose how the CRISPR machinery would functionally link to the transposition machinery — the molecules responsible for gene ‘jumping’ — and the study proved their hypotheses were correct. As noted by Asst Prof Sternberg, “We showed in our first study how to leverage INTEGRATE for targeted DNA insertions in bacterial cells. These new images, a wonderful collaboration with Israel Fernández’s lab, explain the biology with incredible molecular detail and will help us improve the system by guiding protein engineering efforts.”
“Visualising biology on this scale is truly amazing and can easily excite even those unfamiliar with the topic,” added Columbia PhD student Tyler Halpin-Healy, first author of the study. “The quality of this work, and the speed at which it was accomplished, is emblematic of the collaborative environment afforded by great mentors like Sam and Israel.”
In addition to informing future engineering efforts, the structures highlight a possible proofreading checkpoint. Existing CRISPR technologies often suffer from so-called ‘off-target effects’, in which unintended sequences are promiscuously modified. The new structures reveal how Cascade and TniQ work together to ensure that only the correct ‘on-target’ sequences are marked for DNA insertion. The researchers plan to further explore this checkpoint while developing the tool for new therapeutic approaches to disease.
Please follow us and share on Twitter and Facebook. You can also subscribe for FREE to our weekly newsletters and bimonthly magazine.
Antibiotics hinder vaccine response in infants
Infants who received antibiotics in the first few weeks of life had significantly lower levels of...
Colossal announces 'de-extinction' of the dire wolf
Colossal Biosciences has announced what it describes as the rebirth of the dire wolf, which would...
Aspirin could prevent some cancers from spreading
The research could lead to the targeted use of aspirin to prevent the spread of susceptible types...




