Counting molecules with a mobile phone
Scientists from the California Institute of Technology have invented a technique that will help bring emerging diagnostic capabilities out of laboratories and to the point of care. Writing in the journal ACS Nano, the researchers document their efforts to make diagnostic health care a reality in areas with limited resources, where the procedures required to detect molecular markers are too complex or expensive to be used outside of a central laboratory.
To address the need for a robust readout system for quantitative diagnostics, researchers from the lab of Professor Rustem Ismagilov lab invented a visual readout method that uses analytical chemistries and image processing to provide unambiguous quantification of single nucleic-acid molecules that can be performed by any camera phone. The readout method is validated using RNA from the hepatitis C virus, HCV RNA.
The work utilises a microfluidic technology called SlipChip, which was invented in the Ismagilov lab several years ago. A SlipChip serves as a portable lab-on-a-chip and can be used to quantify concentrations of single molecules. Each SlipChip encodes a complex program for isolating single molecules (such as DNA or RNA) along with chemical reactants in nanolitre-sized wells.
The chip consists of two plates that move or ‘slip’ relative to one another, with each slip joining or separating the hundreds or even thousands of tiny wells, either bringing reactants and molecules into contact or isolating them. The architecture of the chip enables the user to have complete control over these chemical reactions and can prevent contamination, making it suitable for a user-friendly, robust diagnostic device.
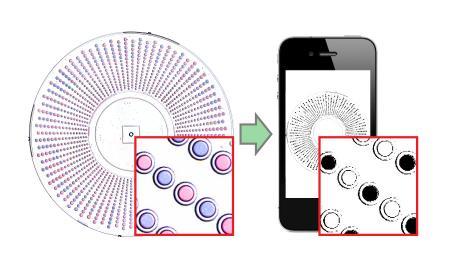
The new visual readout method integrates special indicator chemistries into the wells of the SlipChip device. After an amplification reaction, the well changes colour depending on whether the reaction in it was positive or negative. If a SlipChip is being used to count HCV RNA molecules in a sample, a well containing an RNA molecule that amplified during the reaction would turn blue, whereas a well lacking an RNA molecule would remain purple.
To read the result, a user simply takes a picture of the entire SlipChip using any camera phone. Previous SlipChip technologies utilised a chemical that would fluoresce when a reaction took place within a well, providing readouts which were unfortunately either too subtle for phone camera detection or required specific lighting conditions. However, the new method processes the photo using a ratiometric approach that transforms the colours detected by the camera’s sensor into an unambiguous readout of positives and negatives.
“The readout process we developed can be used with any cell phone camera,” said Jesus Rodriguez-Manzano, one of two first authors on the paper. “It is rapid, automated and doesn’t require counting or visual interpretation, so the results can be read by anyone — even users who are colour blind or working under poor lighting conditions.
“This robustness makes our visual readout method appropriate for integration with devices used in any setting, including at the point of care in limited-resource settings. This is critical because the need for highly sensitive diagnostics is greatest in such regions.”
Non-invasive blood test helps rule out oesophageal cancer
Designed and developed in Australia, the PromarkerEso test is designed to offer a quick,...
Taste-based flu test enables rapid diagnosis
The diagnostic tool consists of the sensor molecule thymol and a virus-specific sugar building...
New international partnerships back Australia's space industry
The Space Industry Association of Australia has commended the Australian Government on some major...







