Imaging CRISPR in action
US scientists have managed to generate near-atomic resolution snapshots of gene editing tool CRISPR, in a breakthrough that provides the structural data necessary for efforts to improve the efficiency and accuracy of CRISPR for biomedical applications.
A subtype of the CRISPR-Cas system, CRISPR-Cas3 is a molecular tool for precision gene editing in biomedical research. But aspects of its mechanism of action — particularly how it searches for its DNA targets — are unclear, and concerns about unintended off-target effects have raised questions about the safety of CRISPR-Cas for treating human diseases.
“To solve problems of specificity, we need to understand every step of CRISPR complex formation,” said Maofu Liao, an assistant professor at Harvard Medical School and co-senior author of the study.
Writing in the journal Cell, Liao and his colleagues describe for the first time the exact chain of events as the CRISPR complex loads target DNA and prepares it for cutting by the Cas3 enzyme. These structures reveal a process with multiple layers of error detection — a molecular redundancy that prevents unintended genomic damage, according to the researchers.
“Our study now shows the precise mechanism for how invading DNA is captured by CRISPR, from initial recognition of target DNA and through a process of conformational changes that make DNA accessible for final cleavage by Cas3,” said Liao.
CRISPR-Cas is an adaptive defence mechanism that bacteria use to fend off viral invaders. This process involves bacteria capturing snippets of viral DNA, which are then integrated into its genome and which produce short RNA sequences known as crRNA (CRISPR RNA). crRNA is then loaded onto members of the CRISPR family of enzymes, which act like sentries that roam the bacteria and monitor for foreign code. If these riboprotein complexes encounter genetic material that matches its crRNA, they chop up that DNA to render it harmless.
CRISPR-Cas subtypes, notably Cas9, can be programmed with synthetic RNA in order to cut genomes at precise locations, allowing researchers to edit genes with unprecedented ease. To better understand how CRISPR-Cas functions, Liao partnered with Ailong Ke, of Cornell University, in a study focusing on type 1 CRISPR — the most common subtype in bacteria.
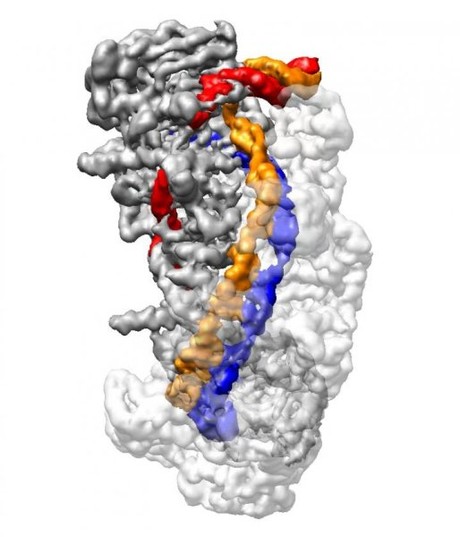
Type 1 CRISPR utilises a riboprotein complex known as CRISPR Cascade for DNA capture and the enzyme Cas3 for cutting foreign DNA. Through a combination of biochemical techniques and cryo-electron microscopy, Liao and Ke reconstituted stable Cascade in different functional states and further generated snapshots of Cascade as it captured and processed DNA at a resolution of up to 3.3 angstroms — or roughly three times the diameter of a carbon atom.
In CRISPR-Cas3, crRNA is loaded onto CRISPR Cascade, which searches for a very short DNA sequence known as PAM that indicates the presence of foreign viral DNA. Liao, Ke and their colleagues discovered that as Cascade detects PAM, it bends DNA at a sharp angle, forcing a small portion of the DNA to unwind. This allows an 11-nucleotide stretch of crRNA to bind with one strand of target DNA, forming a ‘seed bubble’.
The seed bubble acts as a fail-safe mechanism to check whether the target DNA matches the crRNA. If they match correctly, the bubble is enlarged and the remainder of the crRNA binds with its corresponding target DNA, forming what is known as an ‘R-loop’ structure.
Once the R-loop is completely formed, the CRISPR Cascade complex undergoes a conformational change that locks the DNA into place. It also creates a bulge in the second, non-target strand of DNA, which is run through a separate location on the Cascade complex. Only when a full R-loop state is formed does the Cas3 enzyme bind and cut the DNA at the bulge created in the non-target DNA strand. The findings thus reveal an elaborate redundancy to ensure precision and avoid mistakenly chopping up the bacteria’s own DNA.
“To apply CRISPR in human medicine, we must be sure the system is accurate and that it does not target the wrong genes,” said Ke, co-senior author of the study. “Our argument is that the CRISPR-Cas3 subtype has evolved to be a precise system that carries the potential to be a more accurate system to use for gene editing. If there is mistargeting, we know how to manipulate the system because we know the steps involved and where we might need to intervene.”
In contrast to the scalpel-like Cas9, CRISPR-Cas3 acts like a shredder that chews DNA up beyond repair. But while CRISPR-Cas3 currently has limited utility for precision gene editing, it is being developed as a tool to combat antibiotic-resistant strains of bacteria. A better understanding of its mechanisms may broaden the range of potential applications for CRISPR-Cas3.
In addition, all CRISPR-Cas subtypes have been found to utilise some version of an R-loop formation to detect and prepare target DNA for cleavage. The improved structural understanding of this process can now enable researchers to work towards modifying multiple types of CRISPR-Cas systems to improve their accuracy and reduce the chance of off-target effects in biomedical applications.
“Scientists hypothesised that these states existed but they were lacking the visual proof of their existence,” said co-first author Min Luo, a postdoctoral fellow in Liao’s lab. “The main obstacles came from stable biochemical reconstitution of these states and high-resolution structural visualisation. Now, seeing really is believing.”
Droplet microfluidics for single-cell analysis
Discover how droplet microfluidics is revolutionising single-cell analysis and selection in...
PCR alternative offers diagnostic testing in a handheld device
Researchers have developed a diagnostic platform that uses similar techniques to PCR, but within...
Urine test enables non-invasive bladder cancer detection
Researchers have developed a streamlined and simplified DNA-based urine test to improve early...




