Single virus detection techniques for faster disease diagnosis
Two new optical techniques show promise for counting individual viruses outside the laboratory.
To test the severity of a viral infection, clinicians try to gauge how many viruses are packed into a certain volume of blood or other bodily fluid. This measurement, called viral load, helps doctors diagnose or monitor chronic viral diseases such as HIV/AIDS and hepatitis. However, the standard methods used for these tests are only able to estimate the number of viruses in a given volume of fluid. Now two independent teams have developed new optics-based methods for determining the exact viral load of a sample by counting individual virus particles. These new methods are faster and cheaper than standard tests and they offer the potential to conduct the measurements in a medical office or hospital instead of a laboratory.
One research group, led by electrical engineer and bioengineer Aydogan Ozcan of UCLA, is working to directly image single virus particles using holographic microscopy. The other, led by electrical engineer Holger Schmidt of the University of California, Santa Cruz (UCSC), is detecting single particles tagged with fluorescent labels on a microfluidic chip. Both teams expect to use their work to develop commercial instruments useful for on-site diagnosis and monitoring with rapid results and fast turnaround.
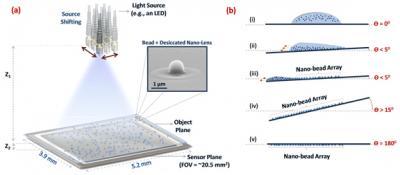
Ozcan’s UCLA team has demonstrated the ability to capture optical images of single viruses and nanoparticles over a comparatively large field of view - about the size of a postage stamp - using nanolenses that self-assemble around the virus particles like little magnifying glasses.
“Because viruses are very small - less than 100 billionths of a metre - compared to the wavelength of light, conventional light microscopy has difficulty producing an image due to weak scattering of sub-wavelength particles,” Ozcan says. When lighted, the team’s new nanolens-nanoparticle assembly projects a hologram that can be recorded using a CMOS imager chip (a type of semiconductor-based light detector) and digitally reconstructed to form an optical image of the particle. “The resulting image improves the field of view of a conventional optical microscope by two orders of magnitude,” says Ozcan.
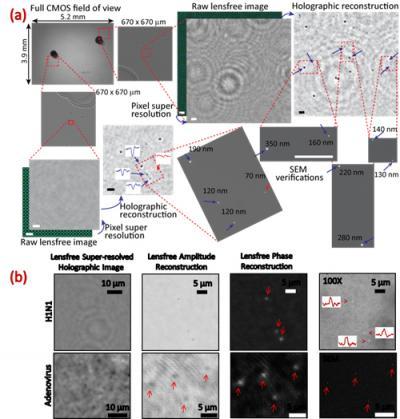
This wide field of view allows the device to form images of many nanoparticles in a single photograph and provides a high-throughput platform for a direct and accurate viral load count. The instrument can be made sufficiently compact and lightweight for field applications and, attached to a cell phone, could become useful even in remote locations.
While Ozcan’s group visually counts individual viruses, Schmidt’s counts them by detecting their nucleic acids - the genetic make-up of the viruses. The nucleic acids are labelled with a fluorescent dye, and light from the fluorescence is detected as they pass through a channel in a microfluidic chip about the size of a thumbnail.
Current tests for determining viral load generally rely on polymerase chain reaction (PCR), which amplifies a small sample of nucleic acid, such as DNA, and makes it easier to detect. “The gold standard for viral load detection is PCR, due to its sensitivity and specificity,” Schmidt says, but PCR is limited to merely estimating the number of viruses. In contrast, the new method counts real particles as they pass through the fluorescence detector on the chip. “We have demonstrated actual virus counts of specific nucleic acids in less than 30 minutes with minimal sample workup,” Schmidt says. So far, the group has collected reliable data on samples diluted to a point well within the range required for clinical detection.
Unlike direct visualisation techniques, Schmidt’s chip-based method requires that the targeted virus particles be labelled. The labelling technique would allow clinicians to target specific viruses while ignoring unlabelled background material. This makes the process potentially useful in situations where clinicians already know what they are looking for - often the case for viral load tests.
The chip is currently housed in an instrument about 30 cm square, making the device portable. Along with rapid analysis turnaround, this portability should make the technique useful for point-of-treatment tests. In addition to detecting viruses, the device may also find uses as a sensor for cancer biomarkers, for environmental analyses of chemicals and even in industrial production monitoring.
Blood test for chronic fatigue syndrome developed
The test addresses the need for a quick and reliable diagnostic for a complex,...
Droplet microfluidics for single-cell analysis
Discover how droplet microfluidics is revolutionising single-cell analysis and selection in...
PCR alternative offers diagnostic testing in a handheld device
Researchers have developed a diagnostic platform that uses similar techniques to PCR, but within...





